Lipid Height Calculations¶
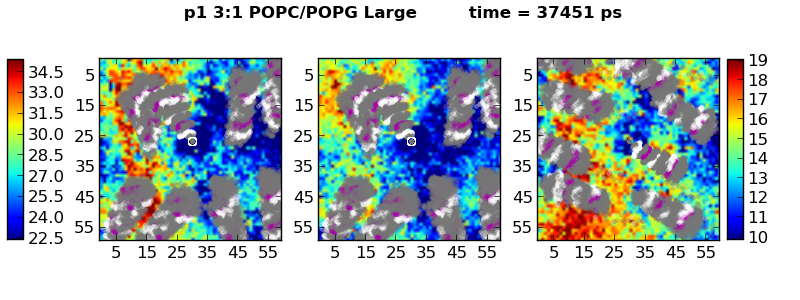
The HeightPlotter code calculates the average height of lipids (Phosphate or C2 to midplane distance) and the bilayer thickness. The output contains three figures: one for the builayer thickness, one for the upper leaflet height, and one for the lower leflet height. The lipids heights are averages over a specified block of simualtion time (trjst to trjsp). If this time is too short, then area on the plots will have a height of zero. If blocks are too short, then peptides may blur as averages flatten.
Minimum Requirements¶
- Calculations require:
- System PSF
- System trajectory files
- matplotlib
- MDAnalysis
Notes¶
- Assumptions:
- Two-component bilayer (for 1 component system, set both LipidA and LipidB to same peptide. Trajectory name is dyn$N.trj, where $N is a number between TrjSt and TrjSp or, if ‘path’ ends with ‘.trj’ or ‘.dcd’, read only that file.
Peptides are colored as gray, purple (Charged), and white (Histidine) circles. This should be easy to modify in furture versions.
Optional Keywords for the Input File¶
| Keyword | Values |
|---|---|
| Title | Title on figures |
| PSF | Location of psf |
| path | Path for trajectory files |
| LipidA | Residue name for lipid A |
| LipidB | Residue name for lipid B (use same as Lipid B no Lipid A) |
| trjst | Number of first trajectory file |
| trjsp | Number of last trajectory file |
| out | Output file name |
| Translate | If True, translate each frame so N-termini are at center |
| Rotate | If True, rotate each frame so N-termmini is at the 6 o’clock Position and the C-termini is at the 12 o’clock position. This is best Used when ‘Translate’ is also True |
| pbcsym | Symmetry of unit cell, P1 or P21 only |
| phosphates | If True, use phosphates to determine lipid height. If False, use C2 of sn-2 chain. |
| plottype | ColorMap, Contour, or Voronoi. Voronoi currenty not working. |
| EmptyCell | If cell has zero population, ‘Zero’ keeps a zero value and ‘Neighbors’ Fills the cell with an average of neighboring cells. If all neighboring Cells are empty, the program will crash! |
| ShowNeighbors | If True, show image peptides. If False, don’t. |
| IncludeHist | If True, include histograms of tilt, depths of insertion, and rotation. All three properties must be calculated beforehand. |
Example Input File:
Title p1 3:1 PC/PG P21
PSF /u/bsp6/Proj/piscidin/DMPC_DMPG_FEP1/SetupPcPgP1_FREE/pcpgp1_p21_mini_free.psf
path /u/bsp6/Proj/piscidin/DMPC_DMPG_FEP1/DynPcPgP1_FREE/
LipidA DMPC
LipidB DMPG
trjst 43
trjsp 62
out run43-62_skip20
Translate True
Rotate True
pbcsym P21
phosphates False
plottype ColorMap
EmptyCell Neighbors
ShowNeighbors False
IncludeHist True